
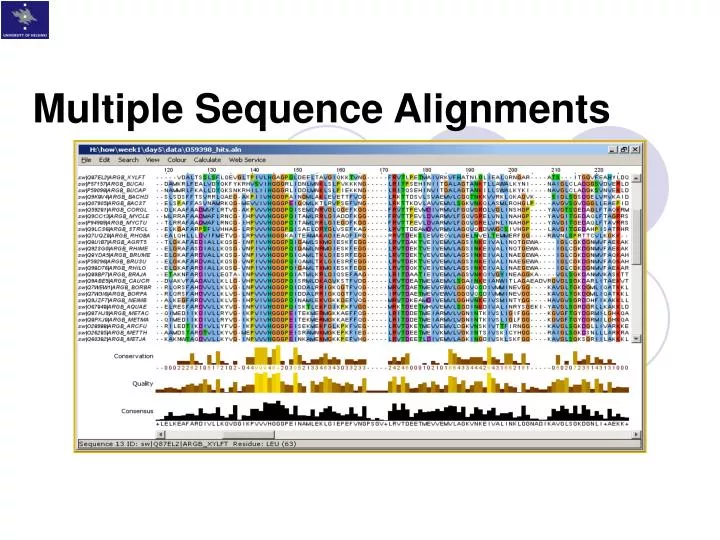
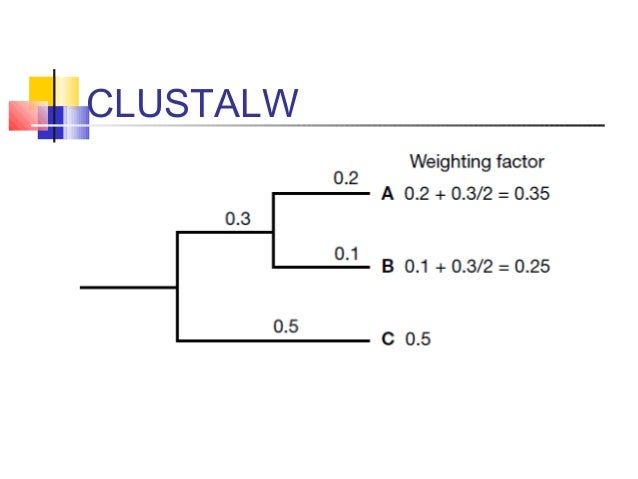
On the BAliBASE benchmark, T-Coffee achieves the best results reported prior to MUSCLE, but has a high time and space complexity that limits the number of sequences it can align to typically around one hundred. A variant of the progressive approach is used by T-Coffee, which builds a library of both local and global alignments of every pair of sequences and uses a library-based score for aligning two profiles. Current progressive algorithms are typically practical for up to a few hundred sequences on desktop computers, the best-known of which is CLUSTALW. If the node is a leaf, the profile is the corresponding sequence otherwise its profile is produced by a pair-wise alignment of the profiles of its child nodes (Figure 2). A profile (a multiple alignment treated as a sequence by regarding each column as a symbol) is then constructed for each node in the binary tree. A more popular strategy is the progressive method (Figure 1), which first estimates a phylogenetic tree.
Stochastic methods such as Gibbs sampling can be used to search for a maximum objective score, but have not been widely adopted. It can be achieved by dynamic programming with time and space complexity O( L N) in the sequence length L and number of sequences N, and is practical only for very small N. A common heuristic is to seek a multiple alignment that maximizes the SP score (the summed alignment score of each sequence pair), which is NP complete. No tractable method for finding an optimal graph is known for biologically realistic models, and simplification is therefore required. This graph makes the history explicit (it can be interpreted as a phylogenetic tree) and implies an alignment. While multiple alignment and phylogenetic tree reconstruction have traditionally been considered separately, the most natural formulation of the computational problem is to define a model of sequence evolution that assigns probabilities to all possible elementary sequence edits and then to seek an optimal directed graph in which edges represents edits and terminal nodes are the observed sequences. We also describe a new method for evaluating objective functions for profile-profile alignment, the iterated step in the MUSCLE algorithm. We introduce a new option designed for high-throughput applications, MUSCLE-fast. Here, we describe the MUSCLE algorithm more fully and analyze its complexity.

We recently introduced MUSCLE, a new MSA program that provides significant improvements in both accuracy and speed, giving only a summary of the algorithm. Obtaining biologically accurate alignments is also a challenge, as the best methods sometimes fail to align readily apparent conserved motifs. Complexity is of increasing relevance due to the rapid growth of sequence databases, which now contain enough representatives of larger protein families to exceed the capacity of most current programs. Two attributes of MSA programs are of primary importance to the user: biological accuracy and computational complexity (i.e., time and memory requirements). Many multiple sequence alignment (MSA) algorithms have been proposed for a recent review, see.
Multiple alignments of protein sequences are important in many applications, including phylogenetic tree estimation, secondary structure prediction and critical residue identification. MUSCLE offers a range of options that provide improved speed and / or alignment accuracy compared with currently available programs. MUSCLE-fast is able to align 1,000 sequences of average length 282 in 21 seconds on a current desktop computer. We find MUSCLE-fast to be the fastest algorithm on all test sets, achieving average alignment accuracy similar to CLUSTALW in times that are typically two to three orders of magnitude less. We test three variants that offer highest accuracy (MUSCLE with default settings), highest speed (MUSCLE-fast), and a carefully chosen compromise between the two (MUSCLE-prog). Accuracy is measured using four benchmarks: BAliBASE, PREFAB, SABmark and SMART. We compare the speed and accuracy of MUSCLE with CLUSTALW, Progressive POA and the MAFFT script FFTNS1, the fastest previously published program known to the author. We also describe a new protocol for evaluating objective functions that align two profiles.
We introduce a new option, MUSCLE-fast, designed for high-throughput applications. Here we present a more complete discussion of the algorithm, describing several previously unpublished techniques that improve biological accuracy and / or computational complexity. In a previous paper, we introduced MUSCLE, a new program for creating multiple alignments of protein sequences, giving a brief summary of the algorithm and showing MUSCLE to achieve the highest scores reported to date on four alignment accuracy benchmarks.


 0 kommentar(er)
0 kommentar(er)
